Beta helix: Difference between revisions
AvocatoBot (talk | contribs) m r2.7.1) (Robot: Adding gl:Hélice beta |
m Bot: link syntax |
||
| (42 intermediate revisions by 30 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:1m8n_Choristoneura_fumiferana.png|thumb|right|200px|Monomeric, left-handed |
[[Image:1m8n_Choristoneura_fumiferana.png|thumb|right|200px|Monomeric, left-handed |
||
[[Image:1ezg_Tenebrio_molitor.png|thumb|350px|Dimeric, right-handed |
[[Image:1ezg_Tenebrio_molitor.png|thumb|350px|Dimeric, right-handed |
||
A '''beta helix''' is a [[protein]] structure formed by the association of parallel [[beta sheet |
A '''beta helix''' is a [[Protein tandem repeats|tandem protein repeat]] structure formed by the association of parallel [[beta sheet]] in a helical pattern with either two<ref>{{cite web|title=CATH database - folds and homologous superfamilies within the beta 2-solenoid architecture.|url=http://www.cathdb.info/cathnode/2.150|website=[[CATH database]]}}</ref> or three<ref>{{cite web|title=CATH database - folds and homologous superfamilies within the beta 3-solenoid architecture.|url=http://www.cathdb.info/cathnode/2.160|archive-url=https://web.archive.org/web/20110726124637/http://www.cathdb.info/cathnode/2.160|archive-date=26 July 2011|url-status=dead|website=[[CATH database]]}}</ref> faces. The beta helix is a type of [[solenoid protein domain]]. The structure is stabilized by inter-strand [[hydrogen bond]]s, [[protein-protein interaction]]s, and sometimes bound metal [[ion]]s. Both left- and right-handed beta helices have been identified. These structures are distinct from [[jelly-roll fold]]s, a different protein structure sometimes known as a "double-stranded beta helix".<ref name="aik_2012">{{cite journal |last1=Aik |first1=WeiShen |last2=McDonough |first2=Michael A |last3=Thalhammer |first3=Armin |last4=Chowdhury |first4=Rasheduzzaman |last5=Schofield |first5=Christopher J |title=Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases |journal=Current Opinion in Structural Biology |date=December 2012 |volume=22 |issue=6 |pages=691–700 |doi=10.1016/j.sbi.2012.10.001|pmid=23142576 }}</ref><ref name="scop">{{cite web |title=Double-stranded beta-helix |url=http://scop.berkeley.edu/sunid=51181 |website=SCOPe |access-date=29 November 2021}}</ref> |
||
| ⚫ | The first beta-helix was observed in the enzyme [[pectate lyase]], which contains a seven-turn helix that reaches 34 Å (3.4 [[nanometer|nm]]) long. The [[P22 phage]] tail spike protein, a component of the P22 [[bacteriophage]], has 13 turns and in its assembled homo[[protein trimer|trimer]] is 200 Å (20 nm) in length. Its interior is close-packed with no central pore and contains both hydrophobic residues and charged residues neutralized by [[salt bridge]]s. |
||
==Two-stranded helices== |
|||
The simplest beta helix contains two "layers" of beta sheets connected by [[glycine]]-rich six-residue loops that invariably contain an [[aspartate]] to bind one [[calcium]] ion per loop. Each layer consists of a nearly-planar series of parallel hydrogen-bonded beta strands and the two layers together enclose a [[hydrophobic]] core. |
|||
Both pectate lyase and [[Phage P22 tailspike protein|P22 tailspike protein]] contain right-handed helices; left-handed versions have been observed in [[enzyme]]s such as [[UDP-N-acetylglucosamine acyltransferase]] and archaeal [[carbonic anhydrase]].<ref name="pmid8665839">{{cite journal |vauthors=Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC |title=A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila |journal=EMBO J. |volume=15 |issue=10 |pages=2323–30 |date=May 1996 |pmid=8665839 |pmc=450161 |doi= 10.1002/j.1460-2075.1996.tb00588.x}}</ref> Other proteins that contain beta helices include the [[antifreeze]] proteins from the beetle ''[[Tenebrio molitor]]'' (right-handed)<ref name="pmid10917536">{{cite journal |vauthors=Liou YC, Tocilj A, Davies PL, Jia Z |title=Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein |journal=Nature |volume=406 |issue=6793 |pages=322–4 |date=July 2000 |pmid=10917536 |doi=10.1038/35018604 |bibcode=2000Natur.406..322L |s2cid=4385352 }}</ref> and from the [[spruce budworm]], ''[[Choristoneura fumiferana]]'' (left-handed),<ref name="pmid12015145">{{cite journal |vauthors=Leinala EK, Davies PL, Jia Z |title=Crystal structure of beta-helical antifreeze protein points to a general ice binding model |journal=Structure |volume=10 |issue=5 |pages=619–27 |date=May 2002 |pmid=12015145 |doi= 10.1016/s0969-2126(02)00745-1|doi-access=free }}</ref> where regularly spaced [[threonine]]s on the |
|||
==Three-stranded helices== |
|||
Three-stranded beta helices form a distorted triangular prism shape in which each face exhibits parallel inter-strand hydrogen bonding. One of the three sheets that form the repeating [[structural motif]] can appear "bent" relative to the other two, which face each other as in the two-stranded helix. Two of the three linking loops between the sheets can be of arbitrary length and can even contain other [[structural domain]]s; the third is restricted to two resides. A characteristic common hexapeptide repeat found in both left- and right-handed helices is the sequence <math>\mathrm{[LIV]-[GAED]-X_{2}-[STAV]-X}</math>. Known three-stranded helices are appreciably longer than their two-stranded counterparts. |
|||
| ⚫ | |||
| ⚫ | The first beta-helix was observed in the enzyme [[pectate lyase]], which contains a seven-turn helix that reaches 34 Å (3.4 [[nanometer|nm]]) long. The [[P22 phage]] |
||
Members of the [[pentapeptide repeat]] family have been shown to possess a quadrilateral beta-helix structure.<ref name="pmid16388575">{{cite journal |vauthors=Vetting MW, Hegde SS, Fajardo JE |title=Pentapeptide repeat proteins |journal=Biochemistry |volume=45 |issue=1 |pages=1–10 |date=January 2006 |pmid=16388575 |pmc=2566302 |doi=10.1021/bi052130w |display-authors=etal}}</ref> |
|||
Both pectate lyase and P22 tailspike protein contain right-handed helices; left-handed versions have been observed in [[enzyme]]s such as [[UDP-N-acetylglucosamine acyltransferase]] and archaeal [[carbonic anhydrase]]. Other proteins that contain beta helices include the [[antifreeze]] proteins from the beetle ''[[Tenebrio molitor]]'' (right-handed) and from the spruce budworm, ''[[Choristoneura fumiferana]]'' (left-handed), where regularly spaced [[threonine]]s on the |
|||
| ⚫ | |||
| ⚫ | |||
{{Reflist}} |
|||
==External links== |
==External links== |
||
| ⚫ | |||
* [http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.c. |
* [https://web.archive.org/web/20120204065931/http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.c.bbg.html SCOP family of left-handed |
||
| ⚫ | |||
* [http://www.cathdb.info/cathnode/2.160 CATH |
* [http://www.cathdb.info/cathnode/2.160 CATH |
||
{{Protein tandem repeats}} |
|||
| ⚫ | |||
*Branden C, Tooze J. (1999). ''Introduction to Protein Structure'' 2nd ed. Garland Publishing: New York, NY. pp 84-6. |
|||
*Dicker IB and Seetharam S. (1992) "What is known about the structure and function of the ''Escherichia coli'' protein FirA?" ''Mol. Microbiol.'', '''6''', 817-823. |
|||
*Kisker C, Schindelin H, Alber BE, Ferry JG and Rees DC. (1996) "A left-handed |
|||
*Liou YC, Tocilj A, Davies PL and Jia Z. (2000) Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein." ''Nature'', '''406''', 322-324. |
|||
*Leinala EK, Davies PL and Jia Z. (2002) "Crystal Structure of |
|||
*Raetz CRH and Roderick SL. (1995) "A Left-Handed Parallel |
|||
*Steinbacher S, Seckler R, Miller S, Steipe B, Huber R and Reinemer P. (1994) "Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer", ''Science'', '''265''', 383-386. (Right-handed) |
|||
*Vaara M. (1992) "Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme", ''FEMS Microbiol. Lett'', '''97''', 249-254. |
|||
*Yoder MD, Keen NT and Jurnak F. (1993) "New domain motif:the structure of pectate lyase C, a secreted plant virulence factor", ''Science'', '''260''', 1503-1507. (Right-handed) |
|||
{{Protein secondary structure}} |
{{Protein secondary structure}} |
||
{{Spirals}} |
|||
[[Category:Protein folds]] |
[[Category:Protein folds]] |
||
[[Category:Protein structural motifs]] |
|||
[[Category:Helices]] |
[[Category:Helices]] |
||
[[es:Hélice beta]] |
|||
[[gl:Hélice beta]] |
|||
[[ja: |
|||
Latest revision as of 10:43, 12 November 2023

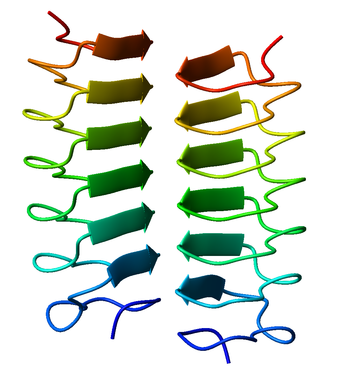
A beta helix is a tandem protein repeat structure formed by the association of parallel beta sheet in a helical pattern with either two[1] or three[2] faces. The beta helix is a type of solenoid protein domain. The structure is stabilized by inter-strand hydrogen bonds, protein-protein interactions, and sometimes bound metal ions. Both left- and right-handed beta helices have been identified. These structures are distinct from jelly-roll folds, a different protein structure sometimes known as a "double-stranded beta helix".[3][4]
The first beta-helix was observed in the enzyme pectate lyase, which contains a seven-turn helix that reaches 34 Å (3.4 nm) long. The P22 phage tail spike protein, a component of the P22 bacteriophage, has 13 turns and in its assembled homotrimer is 200 Å (20 nm) in length. Its interior is close-packed with no central pore and contains both hydrophobic residues and charged residues neutralized by salt bridges.
Both pectate lyase and P22 tailspike protein contain right-handed helices; left-handed versions have been observed in enzymes such as UDP-N-acetylglucosamine acyltransferase and archaeal carbonic anhydrase.[5] Other proteins that contain beta helices include the antifreeze proteins from the beetle Tenebrio molitor (right-handed)[6] and from the spruce budworm, Choristoneura fumiferana (left-handed),[7] where regularly spaced threonines on the
Beta helices can associate with each other effectively, either face-to-face (mating the faces of their triangular prisms) or end-to-end (forming hydrogen bonds). Hence,
Members of the pentapeptide repeat family have been shown to possess a quadrilateral beta-helix structure.[8]
References
[edit]- ^ "CATH database - folds and homologous superfamilies within the beta 2-solenoid architecture". CATH database.
- ^ "CATH database - folds and homologous superfamilies within the beta 3-solenoid architecture". CATH database. Archived from the original on 26 July 2011.
- ^ Aik, WeiShen; McDonough, Michael A; Thalhammer, Armin; Chowdhury, Rasheduzzaman; Schofield, Christopher J (December 2012). "Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases". Current Opinion in Structural Biology. 22 (6): 691–700. doi:10.1016/j.sbi.2012.10.001. PMID 23142576.
- ^ "Double-stranded beta-helix". SCOPe. Retrieved 29 November 2021.
- ^ Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC (May 1996). "A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila". EMBO J. 15 (10): 2323–30. doi:10.1002/j.1460-2075.1996.tb00588.x. PMC 450161. PMID 8665839.
- ^ Liou YC, Tocilj A, Davies PL, Jia Z (July 2000). "Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein". Nature. 406 (6793): 322–4. Bibcode:2000Natur.406..322L. doi:10.1038/35018604. PMID 10917536. S2CID 4385352.
- ^ Leinala EK, Davies PL, Jia Z (May 2002). "Crystal structure of beta-helical antifreeze protein points to a general ice binding model". Structure. 10 (5): 619–27. doi:10.1016/s0969-2126(02)00745-1. PMID 12015145.
- ^ Vetting MW, Hegde SS, Fajardo JE, et al. (January 2006). "Pentapeptide repeat proteins". Biochemistry. 45 (1): 1–10. doi:10.1021/bi052130w. PMC 2566302. PMID 16388575.

