rDNA ITS based identification of Eukaryotes and their communication via DOIs
News, Courses & Workshops, Primer Notes
July 27, 2023: Updated UNITE reference datasets (SH version 9.0) made available on UNITE resources page.
October 17, 2022: New major UNITE SH version (v9.0) released. Reference datasets available on UNITE resources page.
February 2020: New UNITE SH version (v8.2) released. Reference datasets available on UNITE resources page.
In December 2019 NEFOM was funded with 182 000 SEK from SNS and ForBioeconomy for new activities during 2020 under the theme "back to the roots".
October 2018: New article by Nilsson et al.: The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications.
In July 2018 non-Linnaean sequence-based identifiers (UNITE SH codes) were added to GBIF backbone taxonomy (read more from gbif.org).
June, 2017: New UNITE reference datasets (SH version 7.2, minor update) made available on UNITE resources page.
May 4, 2017: Call for Citizen Scientists "Submit specimens for sequencing!"
February 5, 2016: We have set up UNITE Community Twitter account for news and updates. Follow us, and you are up-to-date with UNITE news.
October 13, 2015: New UNITE homepage released.
March 2, 2015: Unified system for the DNA based fungal species (ver. 7.0) released.
October 14, 2013: UNITE homepage updated: Unified system for the DNA based fungal species (ver. 5.0) released.
March-August 2013: New papers relevant to the UNITE effort: Kõljalg et al. 2013, Lindahl et al. 2013, Bentsson-Palme et al. 2013, Lindner et al. 2013, Bates et al. 2013, and Hyde et al. 2013.
September 2012: DNA sequences need quality time too - guidelines for quality control published
Nilsson RH, Tedersoo L, Abarenkov K, Ryberg M, Kristiansson E, Hartmann M, Schoch CL, Nylander JAA, Bergsten J, Porter TM, Jumpponen A, Vaishampayan P, Ovaskainen O, Hallenberg N, Bengtsson-Palme J, Eriksson KM, Larsson K-H, Larsson E, Kõljalg U (2012) Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys 4: 37-62. doi: 10.3897/mycokeys.4.3606
Summer 2010: The proportion of reverse complementary ITS sequences in INSD is growing rapidly, with potentially detrimental effects for the unaware user. We have developed a software solution which we aim to have up and running in UNITE by the end of August 2010.
As per April 2010, PlutoF gives the user the opportunity to extract (locate) ITS1 and ITS2 from (in) fungal ITS sequences. A version for the ribosomal small sub-unit (SSU/16S/18S) is on the way.
As per March 2010, PlutoF now offers generic support for chimera tests of full-length fungal ITS sequences.
In February 2010 the new 454 pipeline for analysis of fungal ITS sequences was added to UNITE.
As per March 2009, support for UNITE ITS sequences from environmental samples (such as soil, ectomycorrhizal roots, orchids etc.) is enabled. You can include sequences with good metadata (like host, locality, habitat, and soil parameters) in your BLAST and galaxieBLAST runs by clicking the 'Envir.' box next to the 'Submit' button. Current number of fungal ITS sequences from environmental samples in UNITE: 7123
As per May 2006, support for INSD (GenBank, EMBL, DDBJ) ITS sequences is enabled. This means that you, in a single BLAST run, can compare your sequences with those in UNITE and INSD. (This pertains to galaxieBLAST as well.) All you have to do is to click the 'INSD' box next to the 'Submit' button; of course, not clicking this button is tantamount to running against UNITE data only. We use emerencia technology (PDF) for the INSD support; all properly annotated INSD fungal ITS sequences should thus be indexed and kept regularly updated (twice a week).
As of November 2005, galaxieBLAST and galaxieHMM support sequences that unintentionally were pasted reverse complementary (?revcomp?). They do this by pattern matching to the first 30 bp of the 5.8S region - unless a sequence features the pattern, the pattern will be looked for in the revcomp of the sequence. If the revcomp of the sequence features this pattern, the analyses will proceed with the revcomp of the sequence. If not, the analyses will proceed with the initial sequence (regardless of the fact that it did not feature the pattern). Thus, partial sequences lacking, say, ITS1 and 5.8S, should also be admissable. We do not anticipate any major complications with this approach - but please inform us of any problems you encounter. However, as explained elsewhere, the galaxie tools are tailored to work on full-length (as opposed to partial) ITS sequences, and you really should use the regular BLAST function of UNITE if you have only a portion of the ITS region available.
April 19-22, 2021: NEFOM workshop "Back to the roots" and linked PhD students course "Metabarcoding Data Management and Analysis" (Online).
Jan 14-15, 2019: PhD students course - Identifying and publishing HTS/Sanger DNA sequence datasets (Copenhagen, Denmark).
May 29 - June 1st, 2017: PhD students course - Metabarcoding Data Management and Open Data (Tartu, Estonia).
April 10-11, 2017: BE Workshop II - Taxonomic annotation of public fungal ITS sequences from the built environment (Aberdeen, UK).
May 23-24, 2016: BE Workshop I - Annotating public fungal ITS sequences from built environment according to the MIxS-Built Environment standard (Gothenburg, Sweden).
September 13-17, 2016: Course "Mycokey - digital key for fungi". Famous Danish mycologist Thomas Læssøe gave a one week course on MycoKey in Tartu, Estonia.
November 2-5, 2016: PhD students course - Biodiversity data management and Open Data (Tartu, Estonia).
January 28-30, 2013: UNITE jamboree - The fungal ITS sequence annotation workshop (Tartu, Estonia).
May 24-26, 2010: PhD students course - Web-based management and analysis of the biodiversity data (Tartu, Estonia).
Identifying ectomycorrhizal fungi - from environmental samples to DNA sequences. A NordForsk research training course 12-24 August 2007.
Primers for amplifying and sequencing fungal rDNA ITS region
PCR primers
To identify ectomycorrhizal (ECM) fungi, ITS1-ITS4, ITS1F-ITS4, and ITS1F-ITS4B are the most widely used primer pairs for PCR. However, using default annealing temperature of 55 degrees centigrade, we have met several difficulties using all of the primer pairs mentioned above.
- ITS1-ITS4 is a pair of universal primers that co-amplifies angiosperm DNA. In case the DNA is extracted from sporocarps, pure cultures, or ECM of conifers, this primer pair amplifies the highest amount of target (ca 600 bp). To our knowledge, ITS1-ITS4 can amplify DNA from nearly all (ECM) fungi.
- ITS1F-ITS4. ITS1F is a fungal specific primer. However, together with ITS4, ITS1F can result in weak amplification of highly concentrated pure angiosperm DNA. We have not observed plant bands on gel (slower) if DNA is extracted from ECM. A problem with ITS1F-ITS4 is gel smearing, that is production of numerous hardly separable bands. Raising annealing temperature a few degrees might resolve this problem. ITS1F tends to result in less target DNA compared to ITS1, but it has proved efficient in amplifying all ECM fungi.
- ITS1F-ITS4B is a pair of strongly basidiomycete-specific primers. In addition to asco-mycetous ECM fungi, Sebacinaceae, Atheliaceae, and some Cortinariaceae species cannot be amplified. We strongly disencourage usage of ITS1F-ITS4B for ECM community studies.
ITS1 or ITS1F together with LR21 appear the most promising primer combination that does not co-amplify plant DNA. These primer pairs amplify nearly all ECM fungi, typically resulting in some 850-900 bp PCR products.
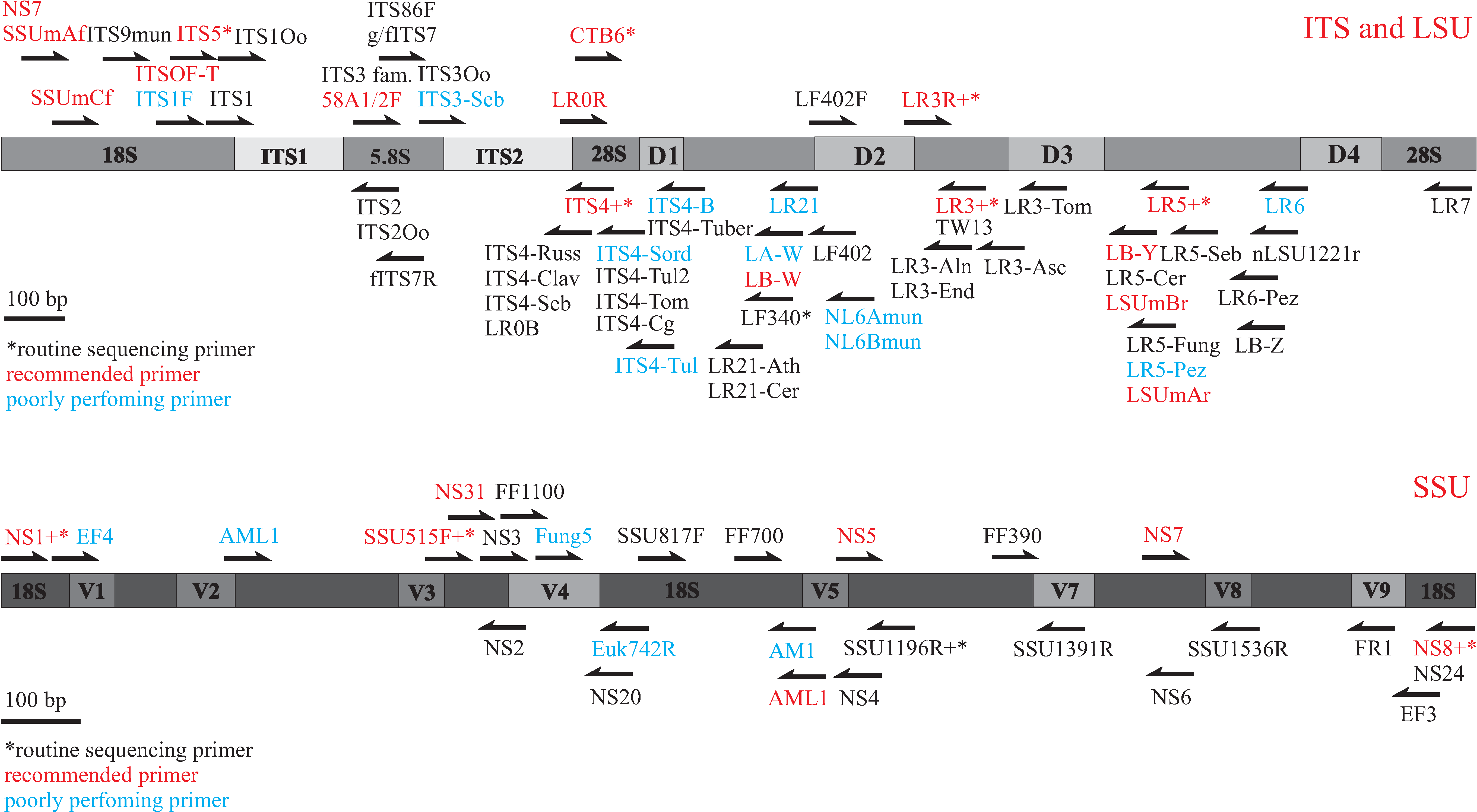
Sequencing primers
In sequencing we have usually applied primers ITS1, ITS2, ITS3, ITS4. ITS2 and ITS3 usually give the best chromatograms. In case of amplification using primer pair ITS1(F)-LR21, ITS3 gives ca 550 bp sequence, covering both ITS2 and ca 200 bp of 28S rDNA. The latter offers more taxonomic opportunities in cases where ITS2 is not matched by other taxa. Sequencing the whole 850 bp with primers ITS1 and LR21 appears to be one of the best solutions.
| Position in gene |
Sequence | Gene/locus | Direction | Target | Remarks | Reference | |
| Universal, fungal primers rDNA | |||||||
| 5.8S_Fungi | CAAGAGATCCGTTGTTGAAAGTT | ITS | rev | Fungi | for paleo-DNA | Epp et al. 2012 | |
| 58A1F | 40 | GCATCGATGAAGAACGC | ITS | fwd | Universal | upgrade of ITS3, excludes Glomeromycota | Martin & Rygiewicz 2005 |
| CTB6 | 39 | GCATATCAATAAGCGGAGG | LSU | fwd | Fungi | for sequencing LR0R products | Garbelotto et al. 1997 |
| ITS1 | 30 | TCCGTAGGTGAACCTGCGG | ITS | fwd | Fungi | excludes Sordariomycetes | White et al. 1990 |
| ITS1F | 90 | CTTGGTCATTTAGAGGAAGTAA | ITS | fwd | Fungi | excludes basal fungi and Tulasnella | Gardes & Bruns 1993 |
| ITS1Fngs | 90 | GGTCATTTAGAGGAAGTAA | ITS | fwd | Fungi | excludes basal fungi and Tulasnella | Tedersoo et al. 2015a,b |
| ITS1ngs | 30 | TCCGTAGGTGAACCTGC | ITS | fwd | Fungi | excludes Sordariomycetes | Tedersoo et al. 2015a,b |
| ITS2 | 40 | GCTGCGTTCTTCATCGATGC | ITS | fwd | Universal | excludes some Agaricales, Tremellales | White et al. 1990 |
| ITS3 | 40 | GCATCGATGAAGAACGCAGC | ITS | fwd | Universal | excludes Cantharellales | White et al. 1990 |
| ITS3mix1-5 | 40 | CANCGATGAAGAACGYRG | ITS | fwd | Universal | excludes Tulasnellaceae | Tedersoo et al. 2014 |
| ITS4 | 40 | TCCTCCGCTTATTGATATGC | ITS | rev | Universal | has central mismatches to some groups and Metazoa and Archaeorhizomycetes | White et al. 1990 |
| ITS4ngs | 40 | TCCTSCGCTTATTGATATGC | ITS | rev | Universal | has central mismatches to some groups and Metazoa and Archaeorhizomycetes | White et al. 1990 |
| ITS5 | 50 | GGAAGTAAAAGTCGTAACAAGG | ITS | fwd | Universal | best sequencing primer for ITSOf/1F products | White et al. 1990 |
| fITS7 | 70 | GTGARTCATCGAATCTTTG | ITS | fwd | Fungi | excludes ca 2% minor groups | Ihrmark et al. 2012 |
| fITS7R | 70 | CAAAGATTCGATGAYTCAC | ITS | rev | Fungi | excludes ca 2% minor groups | L. Tedersoo, unpublished |
| gITS7 | 70 | GTGARTCATCGARTCTTTG | ITS | fwd | Fungi | excludes ca 2% minor groups | Ihrmark et al. 2012 |
| ITS86F | 70 | GTGAATCATCGAATCTTTGAA | ITS | fwd | Fungi, Plants | excludes ca 2% minor groups and Tulasnellaceae | Turenne et al. 1998 |
| fITS9 | 55 | GAACACAGCGAAATGTGA | ITS | fwd | Fungi | excludes ca 4% minor groups | Ihrmark et al. 2012 |
| ITS9mun | 190 | TGTACACACCGCCCGTCG | ITS | fwd | Fungi, Plants | little used | Egger 1995 |
| ITSOF | 90 | ACTTGGTCATTTAGAGGAAGT | ITS | fwd | Fungi | outperforms ITS1F | Tedersoo et al. 2008 |
| LF340 | 336 | TACTTGTKCGCTATCGG | ITS | rev | Universal | for sequencing LB-W products | Tedersoo et al. 2008 |
| LF402 | 402 | TTCCCTTTYARCAATTTCAC | ITS/LSU | rev | Fungi | excludes chytrids, Basidio-yeasts,etc. | Tedersoo et al. 2015a,b |
| LF402F | 402 | GTGAAATTGYTRAAAGGGAA | LSU | fwd | Fungi | excludes chytrids, Basidio-yeasts,etc. | Tedersoo et al. 2015a,b |
| LR0B | 15 | GGTAGTCCTACCTGATTTG | ITS | rev | Basidiomycota | excludes several groups incl Sebacinales | L. Tedersoo, unpublished |
| LR0R | 24 | ACCCGCTGAACTTAAGC | LSU | fwd | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR21 | 370 | ACTTCAAGCGTTTCCCTTT | ITS/LSU | rev | Fungi | excludes many Asco- and Basidiomycota | Hopple & Vilgalys 1994 |
| LR3 | 670 | CCGTGTTTCAAGACGGG | ITS/LSU | rev | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR3R | 670 | GTCTTGAAACACGGACC | LSU | fwd | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR5 | 990 | TCCTGAGGGAAACTTCG | LSU | rev | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR5-Fung | 900 | CGATCGATTTGCACGTCAGA | LSU | rev | Fungi | excludes plants, alveolates and rhizarians, but not stramenopiles or animals | Tedersoo et al. 2008 |
| LR6 | 1165 | CGCCAGTTCTGCTTACC | LSU | rev | Universal | fair performance | Hopple & Vilgalys 1994 |
| LR7 | 1475 | TACTACCACCAAGATCT | LSU | rev | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| nLSU1221r | 1210 | CTAGATGAACYAACACCTT | LSU | rev | Fungi | not enough tested | Schadt et al. 2003 |
| NS7a | 1430 | CAATAACAGGTCTGTGATGC | SSU/ITS | fwd | Universal | widely used | White et al. 1990 (modified) |
| TW13 | 635 | GGTCCGTGTTTCAAGACG | ITS/LSU | rev | Universal | analogue of LR3 | T.J. White, unpublished |
| TW14 | 950 | GCTATCCTGAGGGAAACTTC | LSU | rev | Universal | analogue of LR5 | T.J. White, unpublished |
| Fungal clade-specific primers rDNA | |||||||
| ITS3Seb | 150 | TGAGTGTCATTGTAATCTCAC | ITS | fwd | Sebacinales | excludes ca 20% of target Sebacinales | M.Berbee, unpublished |
| ITS4B | 200 | CAGGAGACTTGTACACGGTCCAG | ITS | rev | Basidiomycota | excludes ca 40% of target taxa | Gardes & Bruns 1993 |
| ITS4B1 | 200 | CAAGRGACTTRTACACGGTCCA | ITS | rev | Basidiomycota | update of ITS4B, still excludes 20% of taxa | Tedersoo et al. 2007 |
| ITS4-Cg | 100 | CACATGGCAARGGCAACCG | ITS | rev | Cenococcum | for improved sequence quality | Bahram et al. 2011 |
| ITS4-Clav | 20 | GGTAGTCCCACCTGATTC | ITS | rev | Clavulina, Cantharellus p.p. | for improved sequence quality | Tedersoo et al. 2011 |
| ITS4-GloeT | 200 | CACAGAAAGCACTTCTCTAA | ITS | rev | Gloeotulasnella | for improved sequence quality | L. Tedersoo, unpublished |
| ITS4-Russ | 20 | AGCGGGTAGTCTCACCC | ITS | rev | Russulaceae | for improved sequence quality | Tedersoo et al. 2011 |
| ITS4-Seb | 20 | TCAGCGGGTARTCCTACTC | ITS | rev | Sebacina clade A | for improved sequence quality | Tedersoo et al. 2011 |
| ITS4-Sord | 100 | CCCGTTCCAGGGAACTC | ITS | rev | Sordariomycetes | for improved sequence quality | Tedersoo et al. 2009 |
| ITS4-Tom | 150 | AACTCGGACGACCAGAGGCA | ITS | rev | Tomentella, Thelephora | excludes 5-10% of Thelephoraceae | Tedersoo et al. 2011 |
| ITS4-Tuber | 230 | CTCGACTCGTAGAAGRCACT | ITS | rev | Tuberaceae | not enough tested | Bonito et al. 2013 |
| ITS4-Tul | 120 | CCGCCAGATTCACACATTGA | ITS | rev | Tulasnellaceae | excludes ca 30% of Tulasnellaceae! | Taylor & McCormick, 2008 |
| ITS4-Tul2 | 50 | TTCTTTTCCTCCGCTGAWTA | ITS | rev | Tulasnella (divergent) | complements to ITS4 and ITS4-Tul for divergent Tulasnellas | Oja et al. 2015 |
| LA-W | 360 | CTTTTCATCTTTCGATCACTC | ITS | rev | Ascomycota | excludes some yeasts, fair PCR product | Tedersoo et al. 2008 |
| LB-W | 360 | CTTTTCATCTTTCCCTCACGG | ITS | rev | Basidiomycota, Zygomycota, Plantae | excludes smuts, some Cantharellus | Tedersoo et al. 2008 |
| LB-wR | 350 | GCGAACAAGTACCGTGAGG | LSU | fwd | Basidiomycota, Zygomycota, Plantae | excludes smuts, some Cantharellus | L. Tedersoo, unpublished |
| LB-y | 870 | TTTGCACGTCAGAATCGCTA | LSU | rev | Basidiomycota (excludes Plantae, other fungi) | good for root tip LSU (excludes some Tulasnella) | Tedersoo et al. 2008 |
| LB-z | 1120 | AAAAATGGCCCACTAGAAACT | LSU | rev | Basidiomycota | good for root tip LSU, excludes some Cantharellus | Tedersoo et al. 2008 |
| LB-Z-Sord | 1020 | GTTTGAGAATGGATGAAGGC | LSU | rev | Sordariomycetes | not enough tested | Tedersoo et al. 2009 |
| LR0R-Tul2 | 40 | CGTTGATTTAAGCATATTAWTC | LSU | fwd | Tulasnella (divergent) | reverse of ITS4-Tul2; not enough tested | L. Tedersoo, unpublished |
| LR21-Ath | 280 | CCAAACAACTCGACTCTTC | ITS | rev | Atheliales | not enough tested | L. Tedersoo, unpublished |
| LR21-Cenoc | 460 | GATGAGCAACATCAGGCAG | ITS | rev | Cenococcum | not enough tested | L. Tedersoo, unpublished |
| LR21-Cer | 300 | CGACTCGTTGAGAGCACAA | ITS | rev | Ceratobasidiaceae | not enough tested | Tedersoo et al. 2011 |
| LR3-Aln | 630 | CCTSAGCACGAACGTGGTA | ITS/LSU | rev | Alnicola, Hebeloma, Entoloma, Inocybe p. parte | for improved sequence quality | L. Tedersoo, unpublished |
| LR3-Asc | 680 | CACYTACTCAAATCCWAGCG | LSU | rev | Ascomycota | not enough tested | Tedersoo et al. 2009 |
| LR3-Cenoc | 580 | TTCAGGCTGGCCGCATTTC | ITS/LSU | rev | Cenococcum | not enough tested | L. Tedersoo, unpublished |
| LR3End | 650 | AYCATTAMGTCAGCGACCTA | LSU | rev | Endogonales | not enough tested | L. Tedersoo, unpublished |
| LR3-Pez | 930 | CMTCRGGATCGGTCGATGG | ITS/LSU | rev | Pezizales | excludes 20% of Pezizales | Tedersoo et al. 2011 |
| LR3-Tul | 570 | GACTCGCATGCAAGGTRCA | ITS/LSU | rev | Tulasnellaceae | excludes 20% of Tulasnellaceae | L. Tedersoo, unpublished |
| LR5-Cer | 860 | CTCTGGCTTCACCCTATG | ITS/LSU | rev | Ceratobasidiaceae | not enough tested | L. Tedersoo, unpublished |
| LR5-Fung | 880 | CGATCGATTTGCACGTCAGA | LSU | rev | Fungi, Metazoa (excl. Plantae) | good for plant tissues | Tedersoo et al. 2008 |
| LR5-Seb | 1000 | ATTCGCTTTACCGCACAAGG | LSU | rev | Sebacinales | not enough tested | Tedersoo et al. 2008 |
| LR5-Tom | 740 | CTACCGTAGAACCGTCTCC | ITS/LSU | rev | Tomentella, Thelephora | excludes 10% of Thelephoraceae | Tedersoo et al. 2008 |
| LR6 Leot-Sord | 1100 | AAAATGGCCCACTAGTGTTG | LSU | rev | Leotiomycetes, SordarioM | not enough tested | Tedersoo et al. 2011 |
| LR6-Asc | 1100 | AAAATGGCCCACTAGTAACG | LSU | rev | Ascomycota, v.a. Leot,SordM | not enough tested | Tedersoo et al. 2011 |
| LR6-Pez | 1100 | CCTCATAAAACRAKATCGTTAC | LSU | rev | Pezizales | not enough tested | L. Tedersoo, unpublished |
| LSUmAr | 930 | [composite] | ITS/LSU | rev | Glomeromycota | good for AMF | Krüger et al. 2009 |
| LSUmBr | 840 | [composite] | ITS/LSU | rev | Glomeromycota | good for AMF | Krüger et al. 2009 |
| NL6Amun | 420 | CAAGTGCTTCCCTTTCAACA | ITS | rev | Ascomycota | not specific, excludes many groups; 1-2 mismatches to Archaeorhizomycetes | Egger 1995 |
| NL6Bmun | 420 | CAAGCGTTTCCCTTTCAACA | ITS | rev | Basidiomycota | not specific, excludes many groups | |
| SSUmAf | 300 | [composite] | ITS | fwd | Glomeromycota | good for AMF | Krüger et al. 2009 |
| SSUmCf | 250 | [composite] | ITS | fwd | Glomeromycota | good for AMF | Krüger et al. 2009 |
| Universal, fungal SSU | |||||||
| FF1100 | 580 | CCAGCTCCAATAGCGTATATTA | SSU | fwd | Fungi | Vainio & Hantola 2000 | |
| FF700 | 980 | GATACCGTIGTAGTCT | SSU | fwd | Fungi | Vainio & Hantola 2000 | |
| FF390 | 1290 | CGATAACGAACGAGACCT | SSU | fwd | Fungi | Vainio & Hantola 2000 | |
| FR1 | 1680 | AICCATTCAATCGGTAIT | SSU | rev | Fungi | Vainio & Hantola 2000 | |
| SSU515f | 500 | GTGCCAGCMGCCGCGGTAA | SSU | fwd | Euakryote/Prokaryote | Cross-domain primer | Turner et al. 1999 |
| SSU515Fngs | 500 | GCCAGCAACCGCGGTAA | SSU | fwd | Euakryote/Prokaryote | Tedersoo et al. 2015a,b | |
| SSU0817f | 790 | TTAGCATGGAATAATRRAATAGGA | SSU | fwd | Eukaryote | Borneman & Hartin 2000 | |
| SSU1196R | 1170 | TCTGGACCTGGTGAGTTTCC | SSU | rev | Eukaryote | Borneman & Hartin 2000 | |
| SSU1196Rngs | 1170 | TCTGGACCTGGTGAGTTT | SSU | rev | Eukaryote | Tedersoo et al. 2015a,b | |
| SSU1391R | 1360 | GACGGGCGGTGWGTRCA | SSU | rev | Eukaryote | Turner et al. 1999 | |
| SSU1536R | 1490 | ATTGCAATGCYCTATCCCCA | SSU | rev | Eukaryote | Borneman & Hartin 2000 | |
| EF4 | 120 | GGAAGGGRTGTATTTATTAG | SSU | fwd | Eukaryote | Smit et al. 1999 | |
| EF3 | 1700 | TCCTCTAAATGACCAAGTTTG | SSU | rev | Eukaryote | Smit et al. 1999 | |
| Fung5 | 680 | GTAAAAGTCCTGGTTCCCC | SSU | rev | Fungi | excludes 10% groups incl. ArchaeorhizoM | Smit et al. 1999 |
| AM1 | 1150 | GTTTCCCGTAAGGCGCCGAA | SSU | rev | Glomeromycota | excludes early diverging groups | Helgason et al. 1998 |
| AML1 | 230 | ATCAACTTTCGATGGTAGGATAGA | SSU | rev | Glomeromycota | Lee et al. 2008 | |
| AML2 | 1170 | GAACCCAAACACTTTGGTTTCC | SSU | rev | Glomeromycota | Lee et al. 2008 | |
| NS1 | 30 | GTAGTCATATGCTTGTCTC | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS2 | 560 | GGCTGCTGGCACCAGACTTGC | SSU | rev | Eukaryote | White et al. 1990 | |
| NS7 | 1400 | GAGGCAATAACAGGTCTGTGATGC | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS6 | 1400 | GCATCACAGACCTGTTATTGCCTC | SSU | rev | Eukaryote | White et al. 1990 | |
| NS3 | 560 | GCAAGTCTGGTGCCAGCAGCC | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS4 | 1120 | CTTCCGTCAATTCCTTTAAG | SSU | rev | Eukaryote | White et al. 1990 | |
| NS5 | 1140 | AACTTAAAGGAATTGACGGAAG | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS8 | 1770 | TCCGCAGGTTCACCTACGGA | SSU | rev | Eukaryote | White et al. 1990 | |
| NS20 | 860 | CGTCCCTATTAATCATTACG | SSU | rev | Eukaryote | White et al. 1990 | |
| NS24 | 1760 | AAACCTTGTTACGACTTTTA | SSU | rev | Eukaryote | White et al. 1990 | |
| NS31 | 535 | TTGGAGGGCAAGTCTGGTGCC | SSU | fwd | Fungi | widely used | Simon et al. 1992 |
| Euk742R | 890 | AAATCCAAGAATTTCACCTCT | SSU | rev | Eukaryote | poor performance | Tedersoo et al. 2015a,b |
| Plant ITS primers | |||||||
| RydinF | 70 | GAACCTTATCRTTTAGAGGAAGG | ITS | fwd | Plantae | ||
| Rydin R | 50 | CCGCCAGATTTTCACGCTGGGC | ITS | rev | Plantae | ||
| ITS4Ang | 20 | GTARTCCCGCCTGACCTG | ITS | rev | Angiospermae | ||
| ITS4PL | 260 | TTCCCAAACAACCCGACTCG | ITS | rev | Plantae | matches many fungi | |
| ITS-OP | 70 | ttaTCATTTAGAGGAAGgAg | ITS | fwd | Plantae | plant analogue of ITS1F | |
| PN16 | 400 | TCCCTTTCAACAATTTCACG | ITS/LSU | rev | Plantae | excludes many groups | |
| 18S-IGS | 20 | ACTACTGGCAGGATCAACCAG | IGS | rev | Plantae, Fungi | ||
| Primers for chloroplast markers | |||||||
| trnL-c | . | CGAAATCGGTAGACGCTACG | trnL | fwd | Plantae | ||
| trnL-d | . | GGGGATAGAGGGACTTGAAC | trnL | rev | Plantae | ||
| trn F | . | ATTTGAACTGGTGACACGAG | trnL | rev | Plantae | ||
| trn E | . | GGTTCAAGTCCCTCTATCCC | trnL | fwd | Plantae | ||
| trn H | . | CGCGCATGGTGGATTCACAATCC | trnL | fwd | Plantae | ||
| psbA | . | GTTATGCATGAACGTAATGCTC | trnL | rev | Plantae | ||
| atpF | . | GAAGTAGTAGGATTGATTCTC | rbcL | rev | Plantae | ||
| rbcL | . | CCCTACAACTCATGAATTAAG | rbcL | fwd | Plantae | ||
| Oomycete primers | |||||||
| ITS1Oo | 0 | GGAAGGATCATTACCACAC | ITS | fwd | Oomycota (99%) | L. Tedersoo, unpublished | |
| ITS2Oo | 40 | GCAGCGKTCTTCATCGRTGT | ITS | fwd | Oomycota (99%) | L. Tedersoo, unpublished | |
| ITS3Oo | 150 | AGTATGYYTGTATCAGTGTC | ITS | fwd | Oomycota (99%) | T. Riit, unpublished | |
| ITS3Perofascia | 140 | CCACCTATGCTACGCTATG | ITS | fwd | T. Riit, unpublished | ||
References
- Bahram M, Põlme S, Kõljalg U, Tedersoo L. 2011. A single European aspen (Populus tremula) tree individual may potentially harbour dozens of Cenococcum geophilum ITS genotypes and hundreds of species of ectomycorrhizal fungi. FEMS Microbiol. Ecol. 75: 313–320.
- Bonito G, Smith ME, Nowak M, Healy RA, Guevara G, Cazares E et al. 2013. Historical biogeography and diversification of truffles in the Tuberaceae and their newly identified Southern Hemisphere sister lineage. PLoS ONE 8: e52765.
- Borneman J, Hartin RJ. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ, Microbiol. 66: 4356–4360.
- Egger KN. 1995. Molecular analysis of ectomycorrhizal fungal communities. Can. J. Bot. 73: S1415-S1422.
- Epp LS, Boessenkool S, Bellemain EP, Haile J, Esposito A, Riaz T, Ers EUs C, Gusarov VI, Edwards ME, Johnsen A, Stenøien H, Hassel K, Kauserud H, Yoccoz N, Brathen KA, Willerslev E, Taberlet P, Coissac E, Brochmann C, 2012. New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems. Molecular Ecology 21: 1821-1833.
- Garbelotto MM, Lee HK, Slaughter G, Popenuck T, Cobb FW, Bruns TD. 1997. Heterokaryosis Is not required for virulence of Heterobasidion annosum. Mycologia 89: 92-102.
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizas and rusts. Mol. Ecol. 2: 113-118.
- Gonzales P, Labarere J. 1998. Sequence and secondary structure of the mitochondrial Small-Subunit rRNA V4, V6, and V9 domains reveal highly species-specific variations within the genus Agrocybe. Appl. Enviro. Microbiol. 64: 4149–4160.
- Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW. 1998. Ploughing up the wood-wide web? Nature 394: 431.
- Hopple JS, Vilgalys R. 1994. Phylogenetic relationships among coprinoid txa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 86: 96-107.
- Ihrmark K, Bödeker ITM, Cruz-Martinez K, Friberg H, Kubartova A, Schenk J, Strid Y, Stenlid J, Brandström-Durling M, Clemmensen KE, Lindahl BD. 2012. New primers to amplify the fungal ITS2 region - evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. , in press.
- Krüger M, Stockinger H, Krüger C, Schüssler A. 2009. DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol. 183: 212-223.
- Lee J, Lee S, Young PW, 2008. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 65: 339-349.
- Martin KJ, Rygiewicz PT. 2005. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5: 28.
- Oja J, Bahram M, Tedersoo L, Kull T, Kõljalg U. 2015. Temporal patterns of orchid mycorrhizal fungi in meadows and forests as revealed by 454 pyrosequencing. New Phytol. 205: 1608–1618.
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84-98.
- Schadt CW, Martin AP, Lipson DA, Schmidt SK. 2003. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301: 1359-1361.
- Simon L, Lalonde M, Bruns TD. 1992. Specific amplification of 18S fungal ribosomal genes from vesicular–arbuscular endomycorrhizal fungi colonising roots. Appl. Environ. Microbiol. 58: 291–295.
- Smit E, Leeflang P, Glandorf B, van Elsas JD, Wernars K. 1999 Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65: 2614–2621.
- Taylor DL, McCormick MK. 2008. Internal transcribed spacer primers and sequences for improved characterization of basidomycetous orchid mycorrhizas. New Phytol. 177: 1020-1033.
- Tedersoo L, Anslan S, Bahram M, Põlme S, Riit T, Kõljalg U, Kisand V, Nilsson RH, Liiv I, Hillebrand F, Abarenkov K. 2015a. Performance of nine primer pair and genetic marker combinations in high-throughput sequencing of fungi. MycoKeys (pending review)
- Tedersoo L, Anslan S, Bahram M, Põlme S, Riit T, Kõljalg U, Nilsson RH, Hillebrand F, Abarenkov K. 2015a. Performance of nine primer pair and genetic marker combinations in high-throughput sequencing of fungi. Science (pending review)
- Tedersoo L, Bahram M, Jairus T, Bechem E, Chinoya S, Mpumba R, Leal M, Randrianjohany E, Razafimandimbison S, Sadam A, Naadel T, Kõljalg U. 2011. Spatial structure and the effects of host and soil environments on communities of ectomycorrhizal fungi in wooded savannas and rain forests of Continental Africa and Madagascar. Mol. Ecol. 20: 3071-3080.
- Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Kõljalg U. 2008. Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol. 180: 479-490.
- Tedersoo L, Pärtel K, Jairus T, Gates G, Põldmaa K, Tamm H. 2009. Ascomycetes associated with ectomycorrhizas: molecular diversity and ecology with particular reference to the Helotiales. Environ. Microbiol. 11: 3166–3178.
- Tedersoo L, Suvi T, Beaver K, Kõljalg U. 2007. Ectomycorrhizal fungi of the Seychelles: diversity patterns and host shifts from the native Vateriopsis seychellarum (Dipterocarpaceae) and Intsia bijuga (Caesalpiniaceae) to the introduced Eucalyptus robusta (Myrtaceae), but not Pinus caribea (Pinaceae). New Phytol. 175: 321-333.
- Turner S, Pryer KM, Miao VPW, Palmer JD. 1999. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Euk. Microbiol. 46: 327–338.
- Vainio EJ, Hantula J. 2000. Direct analysis of wood-inhabiting fungi using denaturing gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104: 927-936.
- White TJ, Bruns TD, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH. (eds). PCR Protocols: A Guide to Methods and Applications. Academic Press: London, pp. 315 – 322.
Alternative online sources
- aftol.org/primers.php
- http://www.clarku.edu/faculty/dhibbett/Protocols_Folder/Primers/Primers.pdf
- biology.duke.edu/fungi/mycolab/primers.htm
- nature.berkeley.edu/brunslab/tour/primers.html
Compiled by Leho Tedersoo
